RRUFF Raman Database Matching
The Rruff Spectral Database and CrystalSleuth
The RRUFF Raman Database can be matched using RRUFF's own CrystalSleuth software.
It will match an unknown against processed RRUFF spectra.
Below is a poor spectrum of Tasmanian crocoite matching to RRUFF crocoites specimens at about 79%.
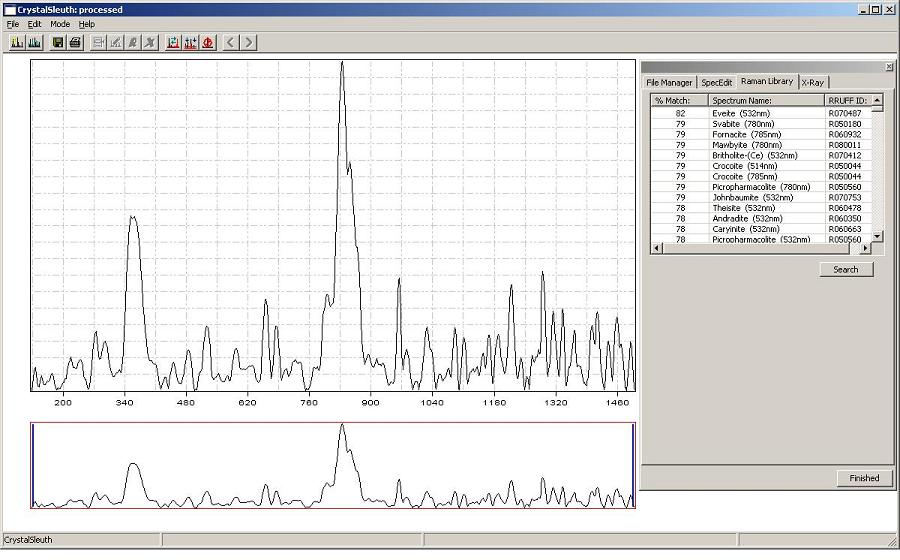
The "processed" spectra in the RRUFF spectral database is wavelength calibrated and
indexed with Raman shift in Kaysers/wavenumbers (cm^-1).
The unknown spectrum's range should be similar (0 to ~1500 cm^-1)
but CrystalSleuth will crop oversize spectra to their
matching regions.
Additionally, the database spectra in most cases seem to be smoothed
and baseline corrected. CrystalSleuth can perform baseline correction
on the unknown spectra.
There are some spectra in the database that have vague profiles
that would be difficult to match. For example, Turquoise.
There are also multiple spectra for some species, sometimes with varying degrees of quality.
There are a few non-mineral spectra as probably useful as reference standards.
CrystalSleuth's Raman Spectra Matching Algoritm/s
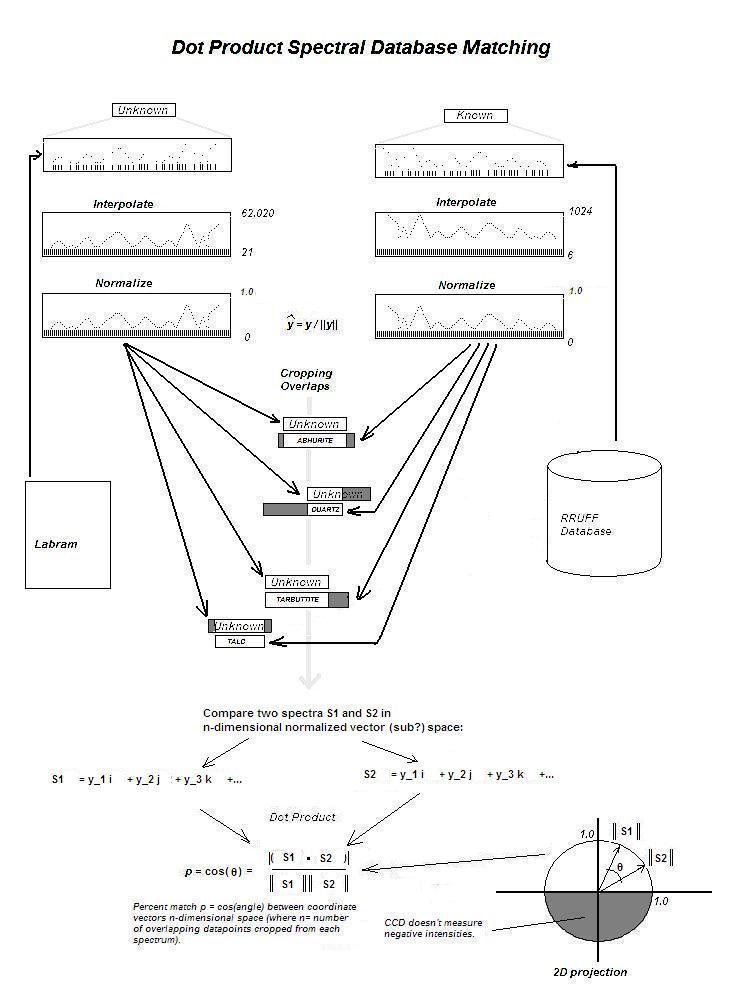
My simplistic description of CrystalSleuth's matching algorithm is
that it interpolates new y-values for equally spaced x-values along the
same x-range. The Y-values are then normalized.
The unknown spectra and each database spectra are cropped and aligned along their
overlapping region. This is followed by a dot product yielding a
percentage of match:
CrystalSleuth seems to be equipped to do a bit more.
It appears that it may be able to implement wavelets
using Haar functions as basis vectors in an orthonormal space.
I'm not sure if it is actually implemented.
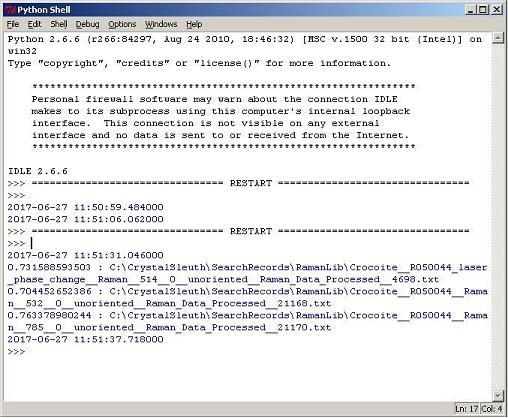
A Python Implementation of CrystalSlueth's (basic) Matching Algorithm
Below, the percent matches of the same specimen to crocoites in the RRUFF library
were produced by our Python program trying to exactly replicate CrystalSleuth's
basic method of matching (without preprocessing and wavelets).
On a Pentium 4 XP sp3 PC it currently takes about 19 minutes with Python
to do a full database match using separate spectra files. CrystalSleuth takes
only about 1 minute using their condensed and pre-processed single file database.
The unknown crocoite spectra was acquired with the Science-Surplus spectrometer
behind the Labram's Raman filter. This was because I'm still having
trouble aligning the Labram's spectrograph with the output from
the microscope. The Science-Surplus spectrometer was calibrated
with Raman shift for the 633nm. The data was saved as a
comma-delimited .rruff file and then smoothed and baseline corrected
with S-G and ALS. Then it was loaded into CrystalSleuth
where it was baseline-corrected (again). The result was this file:
processed.rruff See the raw plot here.
It turns out to be pretty easy to launch CrystalSleuth from our
Python program and feed it a spectrum, so that's probably what
we did instead of reinventing the wheel. We were also able to call
its baseline correction directly from python.
After performing a database match in CrystalSleuth you can
click on any of the matched specimens to see their spectra plotted
along with your unknown's spectrum for visual comparison.
CrystalSleuth is a good program and easy to use but it's basic
dot-product matching is not always reasonable.
Machine Learning and other Artificial Intelligence techniques
are likely to improve on spectra database searches in general.
The Rruff Spectral Database
At this time there are 1,697 unique species in the CrystalSleuth
Raman library represented by a total of 5,129 spectra.
62 spectra have RRUFFIDs that begin with "X".
| Laser | Spectra |
| 514 | 471 |
| 532 | 2464 |
| 780 | 992 |
| 785 | 1202 |
The exciting laser's wavelength, the spectrograph's slit size,
the diffraction grating's rulings, the CCD detector's pixels,...
all have an effect on the resolution of the spectrum.
Specimen and environmental qualities also affect the spectrum.
Phase. Stess. Structural allotropes. Temperature. Chemical purity.
Orientation. etc. ... all influence the spectrum.
How much of the absolute vs. relative peak intensities can be
ignored for the purpose of identification purposes?
Peaks need to be high enough to distinguish them from
background noise and narrow enough to distinguish themselves
from fluorescence and other broader sources of "noise".
What appear to be individual peaks may actually be be overlapping peaks.
If the specimen is pure enough, if the peak resolution is narrow enough,
and if the wavelength scale is well-calibrated,... the peaks should be very
characteristic of the specimen's composition and orientation.
Spectral Line Shape
IR and Raman Spectral Line Shapes
What causes peak broadening?
Temperature,
Sample inhomogeneities; amorphous, other structures
Over-saturation of CCD pixels (spill over)?
Slew rate of ADC
What can cause a peak center to shift or broaden/narrow its bandwidth?
Can a shift affect all peaks or can it affect just one of the characteristic peaks?
Specific vibrational modes can be shifted individually.
A stress on a carbon nanotube in a specific direction can be detected by the
shift of just one of the several peaks in its Raman spectrum.
What causes asymmetric peak broadening?
Incorrect alignment of spectrograph entrance slit with binned CCD?
Examples of two sphalerite spectra from RRUFF (X050150 and R050237):






